🪗 Community Round-Up #16: August 2023
A new platform for the community, re-capping the top 10 M2D2 and LoGG reading groups by all-time views, a genome-wide atlas of human cell morphology, and more...
Hi everyone 👋
Welcome to another issue of the community newsletter! Before we dive into the content, we have a few updates to share:
Although we’re on summer break, we’ve been working hard to bring a brand new experience to our community members. 👀 We’re building the platform that will become the home for the growing communities in AI for drug discovery.
We are looking to get feedback from early users. If you’re interested in helping us make this product the best it can be, please book a time with us for a quick demo/overview!---
If you enjoy this newsletter, we’d appreciate it if you could forward it to some of your friends! 🙏
Let’s jump right in!👇
🧵 Community Twitter Highlights
Here are some of the community’s favourite tweets. Do you have any specific opinions on these threads? Agree or disagree?
Join our Slack community to discuss these topics further!
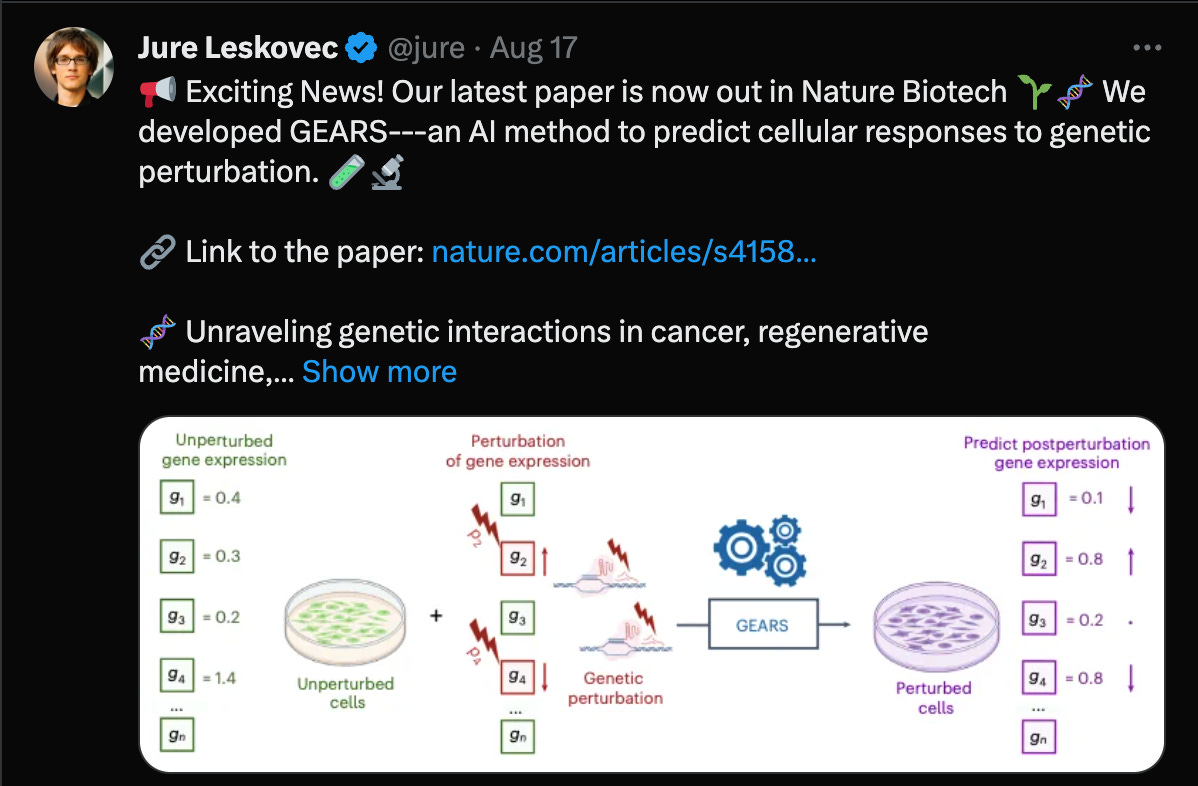
Can deep learning models predict novel multi-gene perturbation outcomes?
A genome-wide atlas of human cell morphology?
Annnnnnd there are way too many tweets for us to show here but check out #ACSFall2023 in San Francisco followed by the #Accelerate23 conference in Toronto on X/Twitter.
📚 Community Reads
Learn more about what the community has been reading!
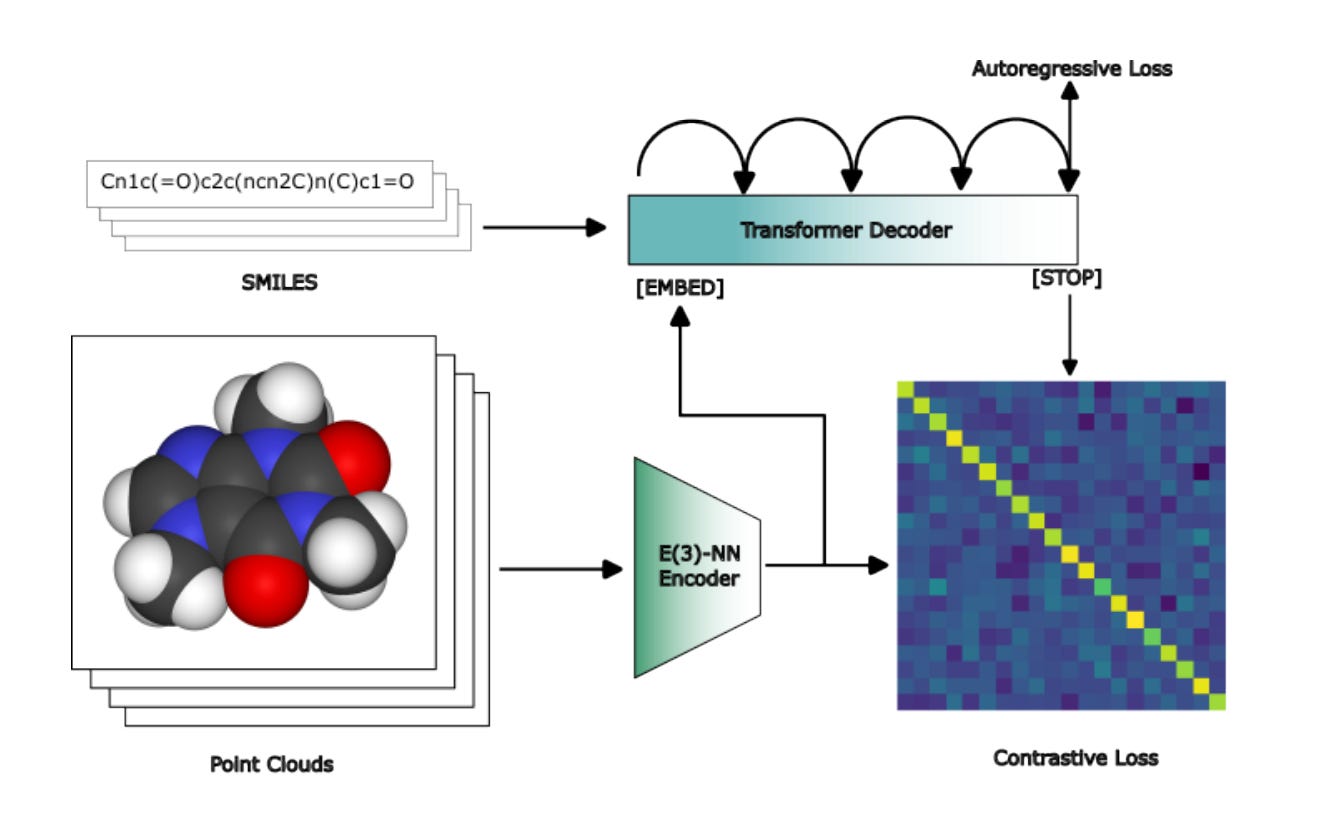
COATI: Multi-Modal Contrastive Pre-Training for Representing and Traversing Chemical Space
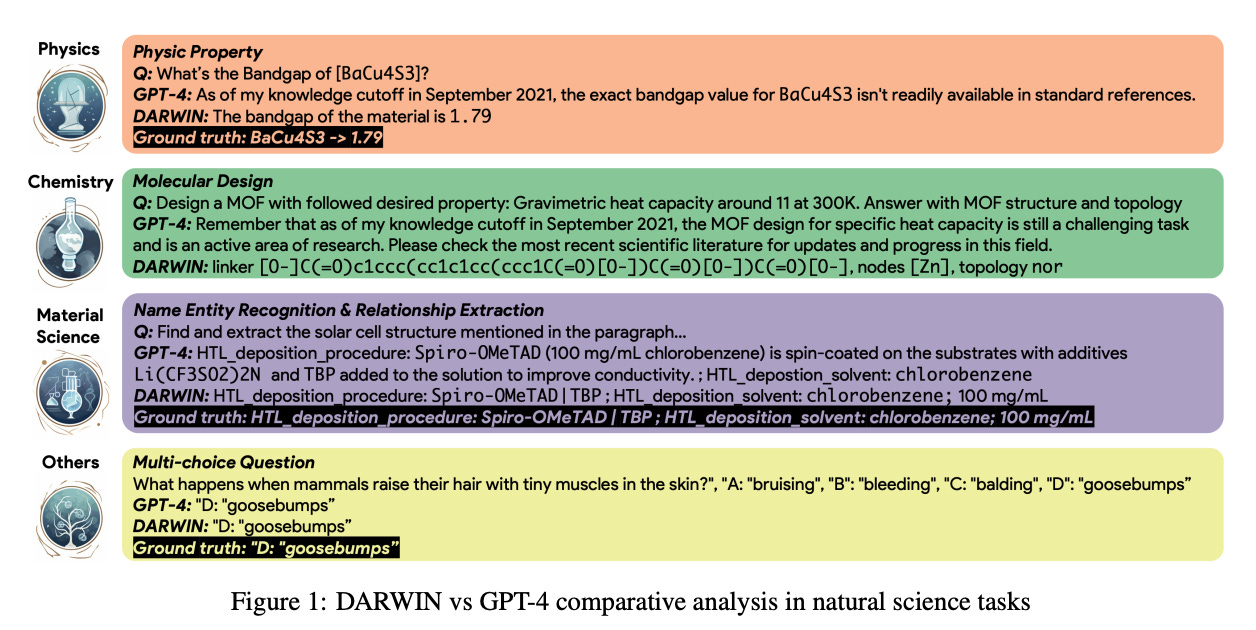
DARWIN Series: Domain-Specific Large Language Models for Natural Science
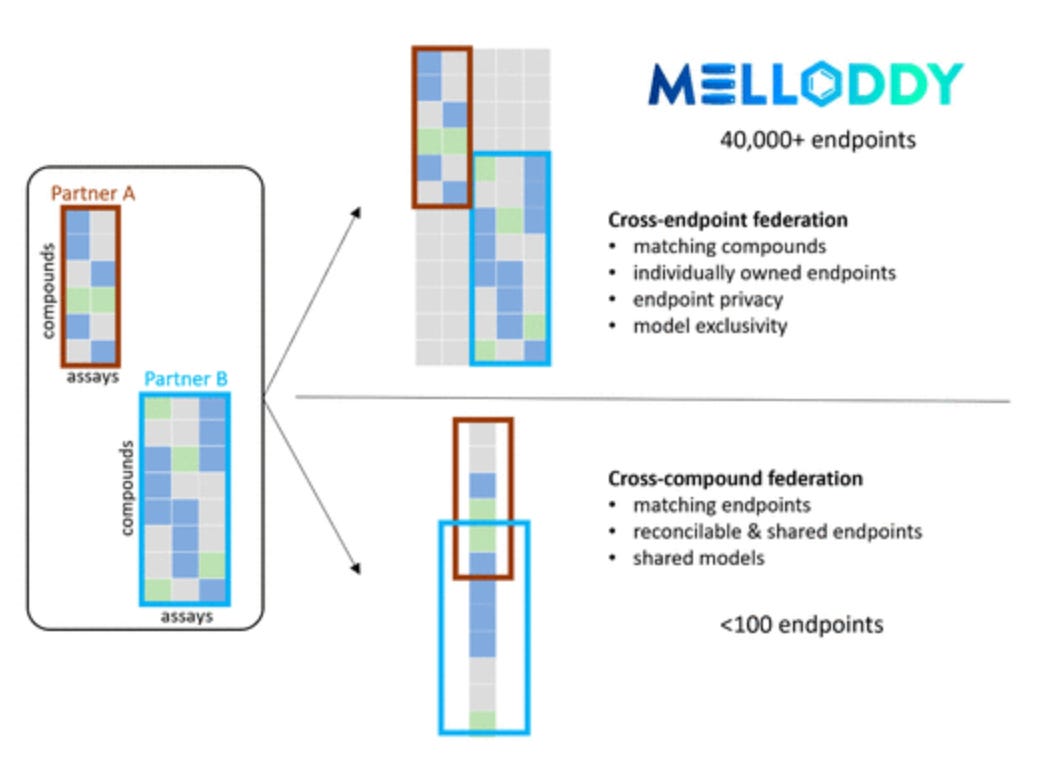
MELLODDY: Cross-Pharma Federated Learning at Unprecedented Scale Unlocks Benefits in QSAR without Compromising Proprietary Information
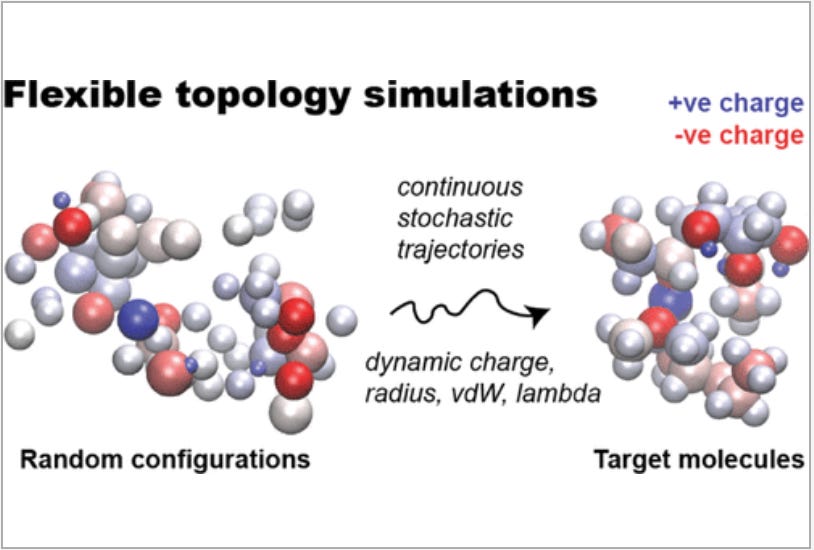
Flexible Topology: A Dynamic Model of a Continuous Chemical Space
🔦 Monthly Spotlight: Tools + Shortcuts
Darwin is an open-source project dedicated to fine-tuning the LLaMA model on scientific literature and datasets. Specifically designed for the scientific domain with an emphasis on materials science, chemistry, and physics, Darwin integrates structured and unstructured scientific knowledge to enhance the efficacy of language models in scientific research.
Check them out on GitHub.
🗃️ In Case You Missed It
Although we’re on summer break, we have a catalogue of nearly 150 videos from our prior recordings from both the M2D2 and LoGG reading groups. You can watch these to your heart’s content on YouTube. 😄
Many people often ask us which video to start with. Here are the top 10 by all-time views on YouTube:
1️⃣ LoGG: Relating Graph Neural Networks to Structural Causal Model by Matej Zečević
2️⃣ M2D2: DiffDock - Diffusion Steps, Twists, and Turns for Molecular Docking by Hannes Stärk, Gabriele Corso, and Bowen Jing
3️⃣ LoGG: Graph Neural Networks and Diffusion PDEs by Ben Chamberlain and James Rowbottom
4️⃣ M2D2: Machine Learning for Scientific Discovery by Yoshua Bengio
5️⃣ M2D2: AlphaFold2, OpenFold, Protein Language Models and Beyond by Nazim Bouatta
6️⃣ M2D2: Converging Advances to Accelerate Molecular Simulation by Max Welling
7️⃣ LoGG: Recent Advances in Deep Learning for Routing Problems by Chaitanya K. Joshi
8️⃣ LoGG: Pure Transformers are Powerful Graph Learners by Jinwoo Kim
9️⃣ M2D2: Euclidean Deep Learning Models for 3D Structures & Interactions of Molecules by the late Octavian-Eugen Ganea
🔟 M2D2: Equivariant 3D-Conditional Diffusion Models for Molecular Linker Design by Ilia Igashov
The M2D2 community lives on Slack with 1,500+ students, professors, researchers and engineers. We aim to demystify the field of AI for drug discovery and merge the vibrant AI & drug discovery communities together to spark new perspectives, provoke discussions, and offer a safe space to share new ideas.
See you at the next issue! 👋