🪗 Community Round-Up: Week of October 9th, 2023
M2D2 Talks are BACK! Also: multi-modal learning, RoseTTAFold All-Atom, LLMs for science, and many more papers
Hi everyone 👋
Welcome to another issue of the community newsletter! With so much happening in the field of AI for drug discovery, we’re now increasing the cadence of the newsletter to once a week. For better organization, we’ve done our best to categorize papers into key topics: ML for Small Molecules, ML for Atomistic Simulations, Multi-Modal Learning, ML for Proteins, LLMs for Science, and Open Source.
We’re also excited to announce that the M2D2 Talks are back! 🚀 We’ll be joined by Prof. Amir Barati Farimani from Carnegie Mellon University who will discuss his recent work in large language molecular representation & learning.
Join us live on Zoom on Tuesday, October 17th at 11 am ET. Find more details here.
If you enjoy this newsletter, we’d appreciate it if you could forward it to some of your friends! 🙏
Let’s jump right in!👇
📚 Community Reads
ML for Small Molecules
Structured State-Space Sequence Models for De Novo Drug Design
Newly proposed S4 models can be formulated as convolutions on SMILES strings during training and can recurrently produce SMILES strings during generation. This study compares S4 with two state-of-the-art approaches: Long Short-Term Memory networks (LSTM) and Generative Pretrained Transformers (GPT) on tasks relevant to drug discovery, with a focus on de novo molecule generation.
ML for Atomistic Simulations
Grad DFT: A Software Library for Machine Learning Enhanced Density Functional Theory
Density functional theory (DFT) is a computational quantum mechanical modeling method used to investigate the electronic structure of atoms and molecules. Machine learning techniques are starting to be explored for improving DFT. This work presents a JAX-based DFT library to enable quick prototyping and experimentation. They also compile a curated dataset of experimental dissociation energies of dimers to support training and benchmarking efforts.
Multi-Modal Learning
Meta-Transformer: A Unified Framework for Multimodal Learning
Unified Multimodal Learning. Meta-Transformer utilizes the same backbone to encode natural language, image, point cloud, audio, video, infrared, hyperspectral, X-ray, time-series, tabular, Inertial Measurement Unit (IMU), and graph data. Meta-Transformer indicates a promising future for developing unified multimodal intelligence with transformers.
NExT-GPT: Any-to-Any Multimodal LLM
An end-to-end general-purpose any-to-any MM-LLM system, NExT-GPT, based on an LLM with multimodal adaptors and different diffusion decoders. NExT-GPT can perceive inputs and generate outputs in arbitrary combinations of text, images, videos, and audio, using existing well-trained highly-performing encoders and decoders. The model is tuned with only a small amount of parameter (1%) of certain projection layers, which not only benefits low-cost training but also facilitates convenient expansion to more potential modalities.
ML for Proteins
Generalized Biomolecular Modeling and Design with RoseTTAFold All-Atom
Models like AlphaFold2 and RoseTTAFold have been impactful for structural biology and caused a lot of excitement, but they don’t model ‘non-protein’ parts. This work presents a RoseTTAFold capable of modelling proteins along with the non-protein components like nucleic acids, metals, small molecules and covalent modifications. The authors also fine-tune on diffusive denoising tasks and can generate binding pockets and proteins that bind to molecules of interest, and they validate the results of these designs in vitro.
SaProt: Protein Language Modeling with Structure-Aware Vocabulary
Large-scale protein language models (PLMs) have achieved remarkable performance on tasks related to protein structure and function, but are limited by explicit consideration for protein structure information. This work introduces a “structure-aware vocabulary”, integrating residue tokens with structure tokens. The structure tokens are derived by encoding the 3D structure of proteins using Foldseek. SaProt is trained on 40 million protein sequences and structures and achieves significant results across 10 downstream tasks. The code and model are available at https://github.com/westlake-repl/SaProt.
Improving Protein Expression, Stability, and Function with ProteinMPNN
Producing natural proteins in biotech is sometimes challenging because we’re using different systems to produce them. Problems such as poor expression, limited solubility and temperature sensitivity can arise. This work uses the deep neural network ProteinMPNN with evolutionary and structural information to increase protein expression, stability and function. They support their predictions with experimental results for two proteins, myoglobin and TEV protease. This approach could be broadly useful for protein expression in biotech.
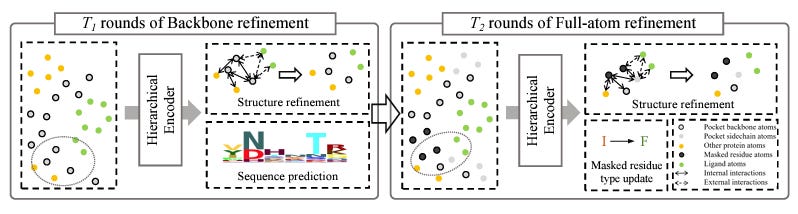
Full-Atom Protein Pocket Design via Iterative Refinement
When designing new proteins to bind specific ligand molecules, properly capturing the protein pocket is vital. FAIR (Full-Atom Iterative Refinement) is designed to address inefficient generation, inadequate context modeling of ligand, and the inability to generate side-chain atoms. In the initial stage, the residue types and backbone coordinates are refined using a hierarchical context encoder, complemented by two structure refinement modules that capture both inter-residue and pocket-ligand interactions. The subsequent stage delves deeper, modeling the side-chain atoms of the pockets and updating residue types to ensure sequence-structure congruence. FAIR is available at https://github.com/zaixizhang/FAIR.
SE(3)-Stochastic Flow Matching for Protein Backbone Generation
Designing new protein structures is a hot topic. This work presents FoldFlow, a series of generative models based on flow-matching over 3D rigid motions (the SE(3) group). These models are more stable and faster to train than diffusion-based approaches, and can map from invariant source distribution to target distribution over SE(3).
LLMs for Science
A Proposed Test for Human-Level Intelligence in AI
In light of the advancing sophistication of AI systems, the need for a robust intelligence test has grown increasingly urgent. While various approaches have been proposed, such as puzzle-solving, natural language comprehension, and adaptive learning, the Eagleman suggests that instead of conventional methods like the Turing or Lovelace tests true intelligence is reflected in an AI's capacity for scientific discovery. The paper introduces two distinct levels of scientific discovery: Level 1, involving the synthesis of scattered scientific facts, and Level 2, requiring the creation of entirely new models and frameworks. Ultimately, Eagleman advocates for using Level 2 discoveries as the benchmark to demonstrate human-level intelligence in AI.
Lost in the Middle: How Language Models Use Long Contexts
The study examines how well language models handle longer input contexts and their performance in tasks like multi-document question answering and key-value retrieval. The authors show that models perform best when relevant information is at the beginning or end of the input, but their performance drops significantly when accessing information in the middle of lengthy contexts. Even explicitly designed long-context models experience decreased performance with longer input.
Large Language Models Encode Clinical Knowledge
Med-PaLM paper from Google/DeepdMind that evaluates the performance of PaLM and its specialized variant, Flan-PaLM, on MultiMedQA datasets. Flan-PaLM achieves remarkable state-of-the-art accuracy across all datasets, notably surpassing the prior state-of-the-art on MedQA by over 17%. However, human evaluation reveals limitations in Flan-PaLM's responses.
To address these gaps, DeepMind introduces instruction prompt tuning, resulting in Med-PaLM. While Med-PaLM shows promise, it remains inferior to clinical expertise. The study highlights improvements in comprehension, knowledge recall, and medical reasoning with model scale and instruction prompt tuning, showcasing LLMs' potential in medicine. However, it underscores the need for robust evaluation frameworks and ongoing method development for safe and helpful LLM models in clinical applications.
ML for Omics
With all of the data being generated in biology, it’s natural to think of leveraging them by training foundation models. Foundation models are large machine learning models trained on a vast quantity of data, with the ability to be adapted to a range of downstream tasks. The following papers show that a range of efforts are underway to create foundation models for different subdomains of biology.
Large-Scale Foundation Model on Single-cell Transcriptomics
This work trained their 100M parameter scFoundation model on over 50 million human single-cell transcriptomics samples. This effort can serve as a foundation model and be useful for gene expression enhancement, tissue drug response prediction, single-cell drug response classification, and more.
An RNA Foundation Model Enables Discovery of Disease Mechanisms and Candidate Therapeutics
Another foundation model, this time for RNA biology. BigRNA was trained on thousands of genome-matched datasets to predict tissue-specific RNA expression, splicing, microRNA sites, and RNA binding protein specificity from DNA sequence. This model accurately predicted the effects of steric-blocking oligonucleotides (SBOs) on increasing the expression of 4 genes, and on splicing 18 exons across 14 genes including disease-related genes.
GET: A Foundation Model of Transcription Across Human Cell Types
GET is designed to uncover regulatory grammars across 213 human fetal and adult cell types. It was trained using chromatin accessibility data and sequence information and can predict gene expression even in previously unseen cell types. The study also provides catalogues of gene regulation and transcription factor interactions, with cell type specificity.
Open Source
AutoGen: Enabling Next-Generation Large Language Model Applications
Designing and implementing a workflow to get the most out of large language models (LLMs) can be challenging. Automating these workflows can help. AutoGen is a framework for automating LLM workflows that offers customizable and conversable agents. The code can be found at https://github.com/microsoft/autogen.
Think we missed something? Join our community to discuss these topics further!
Valence Portal is the home of the AI for drug discovery community. Join here and stay up to date on the latest research, expand your network, and ask questions. In Portal, you can access everything that the community has to offer in a single location:
M2D2 - a weekly reading group to discuss the latest research in AI for drug discovery
LoGG - a weekly reading group to discuss the latest research in graph learning
Blogs - tutorials and blog posts written by and for the community
Discussions - jump into a topic of interest and share new ideas and perspectives
See you at the next issue! 👋