⚛️ M2D2 Community Round-Up #11: March 2023
Introducing molfeat - an open source hub of molecular featurizers. The newsletter also features better quantum chemical property prediction, zero-shot predictions with GNNs, and more...
Hi everyone 👋
Welcome to another issue of the Molecular Modelling and Drug Discovery (M2D2) community newsletter! Before we dive into the content, we have a few updates to share from the community:
We are excited to announce the release of molfeat, an open source hub of molecular featurizers! 🥳 This project has been in the works for a few months now and was led by a core group of developers from the community. We’d like to thank everyone from this community who supported and shared feedback. We hope this new tool helps streamline your molecular ML workflows - get started with molfeat today ❤️
⭐ Show us some love on GitHub ⭐
🌎 Read more about the launch in our newest community blog 🌎
---
If you enjoy this newsletter, we’d appreciate it if you could forward it to some of your friends! 🙏
Let’s jump right in!👇
🧵 Community Twitter Highlights
Here are some of the community’s favourite tweets. Do you have any specific opinions on these threads? Agree or disagree?
Join our Slack community to discuss these topics further!
Design de-novo proteins from… home?

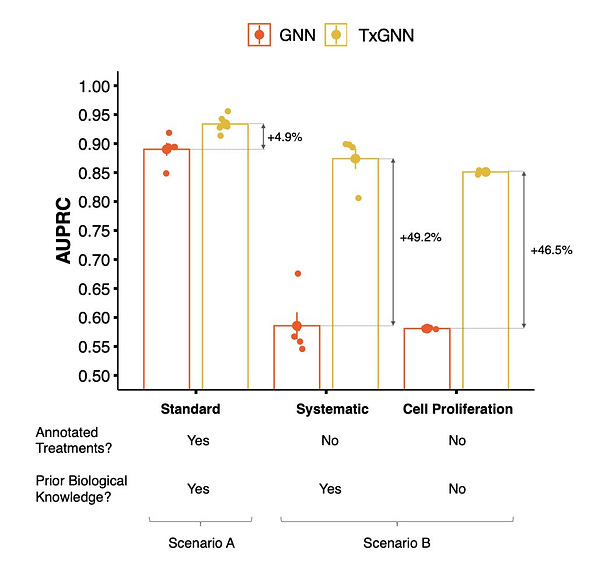
Is there hope for diseases with limited treatment options?

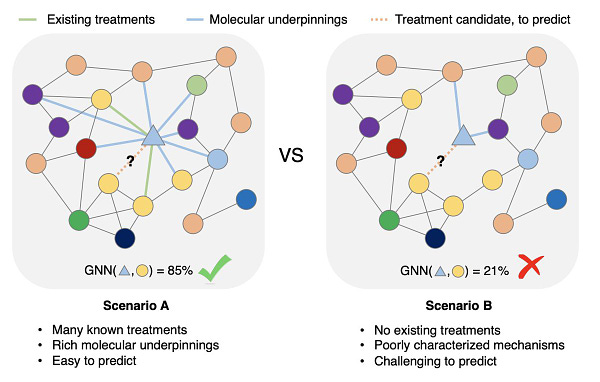

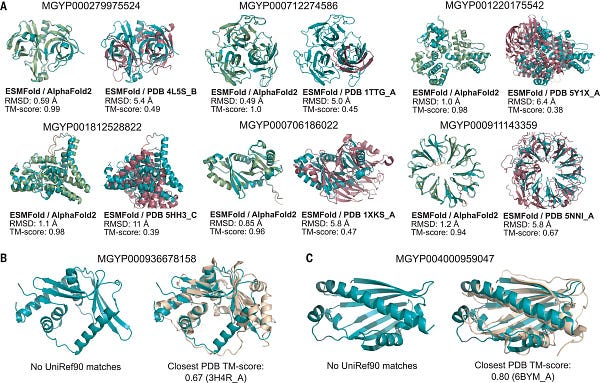
Are we getting closer to solving the data problem?

📚 Community Reads
Learn more about what the community has been reading!
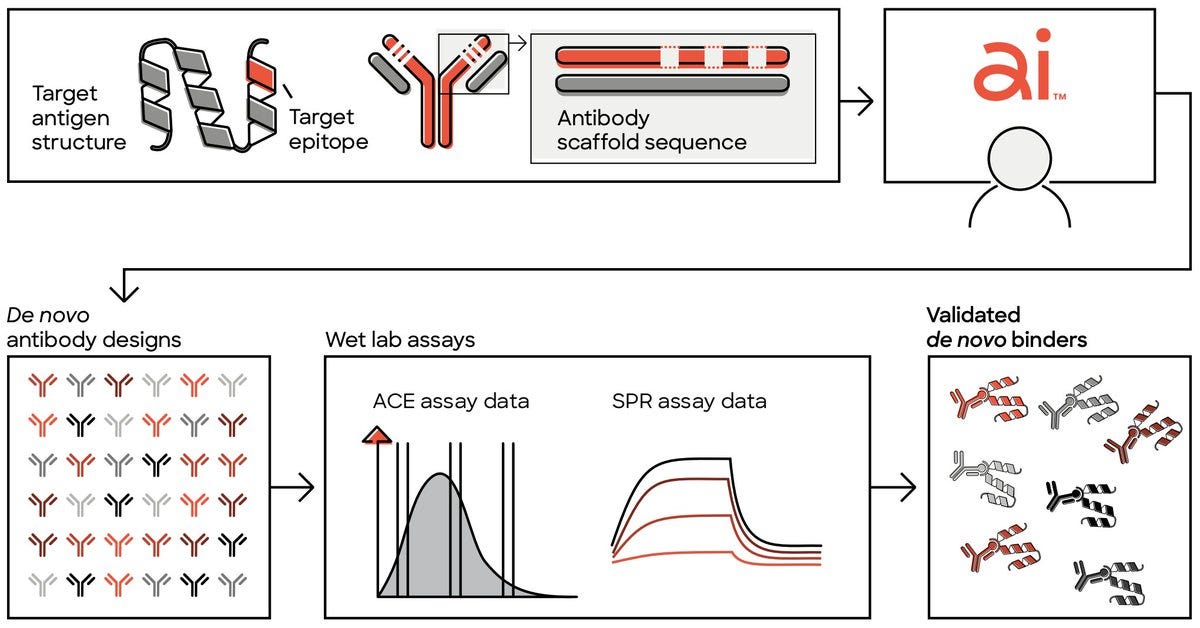
Unlocking De Novo Antibody Design with Generative Artificial Intelligence
ProtFIM: Fill-in-Middle Protein Sequence Design via Protein Language Models
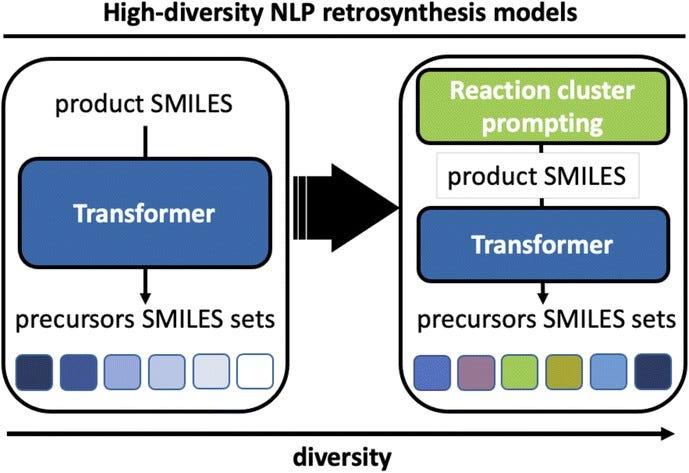
Enhancing Diversity in Language-Based Models for Single-Step Retrosynthesis
A Perspective on Explanations of Molecular Prediction Models
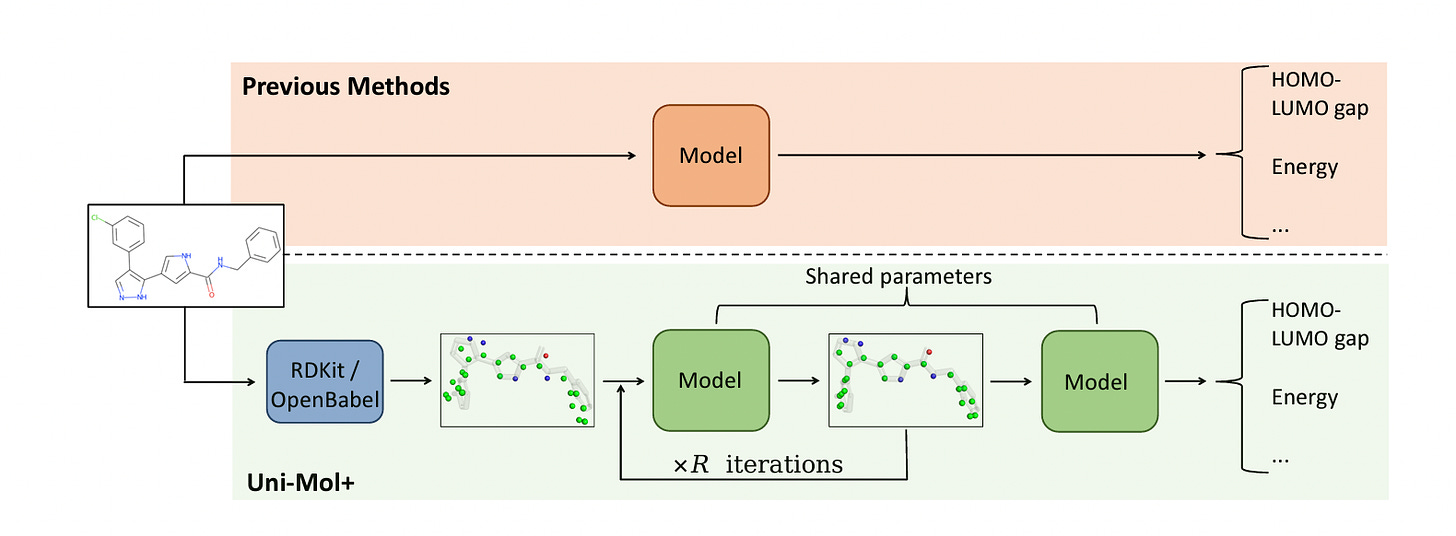
Highly Accurate Quantum Chemical Property Prediction with Uni-Mol+
🔦 Monthly Spotlight: Tools + Shortcuts
Make sure you check out molfeat! molfeat unifies a diverse range of molecular featurizers into a single package. A growing list of the most popular and recent SOTA featurizers are offered, including:
Pre-trained embeddings (e.g. ChemBERTa, ChemGPT and Graphormer, pretrained Graph Isomorphism Networks)
Structural fingerprints (e.g. ECFP and MACCS)
Physico-chemical descriptors (e.g. 2D RDKit descriptors and Mordred)
Our extensive tutorials and documentation make it super easy to get started and contribute something new to the hub. If you find the tool useful, show us some love on GitHub. ⭐
🎭 M2D2 Member Spotlight
This section is dedicated to helping you learn more about the people who are a part of the M2D2 community. If you want to be featured, please message Meilina Reksoprodjo on Slack!
Daniel McNeela
About you: I’m a Ph.D. candidate in Computer Science at the University of Wisconsin, Madison. I also work as a freelance technical and content writer for a number of software and machine learning companies.
Research areas: Geometric deep learning, differential geometry, topological data analysis, and computational drug discovery.
What’s new: Currently working on the OpenDrugDiscovery project.
Looking for: Research collaborations in any of my research areas and potential startup cofounders!
🗃️ In Case You Missed It
Catch up on this month's M2D2 talks here:
Structure-Aware Protein Self-Supervised Learning by Can (Sam) Chen
Interpretable Chirality-Aware GNNs for QSAR Modeling in Drug Discovery by Yunchao (Lance) Liu
Calibration and Generalizability of Probabilistic Models on Low-Data Chemical Datasets by Gary Tom
Fragment-Based Hit Discovery via Unsupervised Learning of Fragment-Protein Complexes by William McCorkindale
⭐ Contributors
Thank you to our Winter newsletter lead, Meilina Reksoprodjo, for putting together this edition of the newsletter. If you’d like to get involved in helping craft future issues, please hit reply or message us on Slack!
The M2D2 community lives on Slack with 900+ students, professors, researchers and engineers. We aim to demystify the field of AI for drug discovery and merge the vibrant AI & drug discovery communities together to spark new perspectives, provoke discussions, and offer a safe space to share new ideas.
See you at the next issue! 👋