☄️M2D2 Community Round-Up #13: April 2023
Releasing the M2D2 community guidelines, LLMs for molecular representations, auto-ML tools for molecular property prediction, and more
Hi everyone 👋
Welcome to another issue of the Molecular Modelling and Drug Discovery (M2D2) community newsletter! Before we dive into the content, we have a few updates to share from the community:
A fun experiment was conducted by the molfeat team - is it necessary to pretrain/finetune LLMs on molecular context to obtain good molecular representations? 🤔
In this experiment, we created the molfeat-hype extension to investigate the performance of embeddings from various LLMs trained without explicit molecular context. molfeat-hype supports two types of LLM embeddings, classic (Llamam, OpenAI, sentence-transformers) and instruction-based (e.g. ChatGPT).
Learn more about the results and add your own plugin to molfeat today!We can’t believe it’s already May ☀️, that means there are only a few more weeks left until the 2023 Molecular Machine Learning conference! It’s happening on May 29th at Mila, there are a few tickets left for those who have yet to register. We are very excited to hear from all of the speakers. 😎
There will also be two poster sessions for researchers to showcase their work! If you’re interested in presenting your work, make sure you submit something ASAP. The deadline is midnight today!We also released the M2D2 community guidelines to help manage and keep our community organized as we continue to grow. Give it a read!
---
If you enjoy this newsletter, we’d appreciate it if you could forward it to some of your friends! 🙏
Let’s jump right in!👇
🧵 Community Twitter Highlights
Here are some of the community’s favourite tweets. Do you have any specific opinions on these threads? Agree or disagree?
Join our Slack community to discuss these topics further!
The next-gen platform for ultra-large virtual screenings?
Can we mimic synthetic experts with ML models? Open positions at the Schwaller group.
Are TikTok breakdowns the future?
📚 Community Reads
Learn more about what the community has been reading!
Uni-Dock: GPU-Accelerated Docking Enables Ultralarge Virtual Screening
Best Practices on QM/MM Simulations of Biological Systems
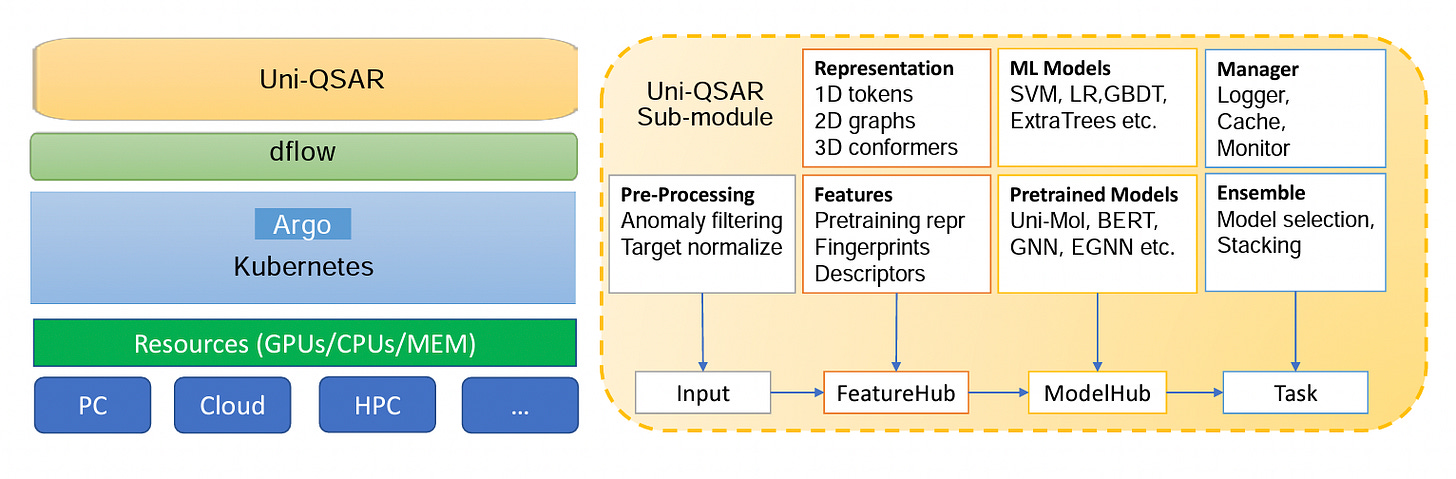
Uni-QSAR: an Auto-ML Tool for Molecular Property Prediction
Molecular Property Prediction by Semantic-invariant Contrastive Learning
PeSTo: Parameter-Free Geometric Deep Learning for Accurate Prediction of Protein Binding Interfaces
Novel Molecular Representations Using Neumann-Cayley Orthogonal Gated Recurrent Unit
DrugormerDTI: Drug Graphormer for Drug–Target Interaction Prediction
Does a Machine-Learned Potential Perform Better Than an Optimally Tuned Traditional Force Field? A Case Study on Fluorohydrins
🔦 Monthly Spotlight: Tools + Shortcuts
M2D2 community member Desmond Gilmour added a new feature into datamol - lasso highlighting! This enhances the visualization process on datamol, users can quickly identify and visualize specific parts or features of a molecule.
This can be useful when identifying functional groups, comparing different molecules, and analyzing molecular structure and properties. Read more about lasso highlighting in Desmond’s blog. 🪢
🗃️ In Case You Missed It
Catch up on this month's M2D2 talks here:
Cell Morphology-Guided De Novo Hit Design by Conditioning GANs on Phenotypic Image Features by Paula A. Marin Zapata
Applying Active Learning in Drug Discovery by Pat Walters & James Thompson
Protein Representation Learning by Geometric Structure Pretraining by Zuobai Zhang
Machine Learning Molecules by Gianni De Fabritiis
⭐ Contributors
Thank you to our Winter newsletter lead, Meilina Reksoprodjo, for putting together this edition of the newsletter. If you’d like to get involved in helping craft future issues, please hit reply or message us on Slack!
The M2D2 community lives on Slack with 1,100+ students, professors, researchers and engineers. We aim to demystify the field of AI for drug discovery and merge the vibrant AI & drug discovery communities together to spark new perspectives, provoke discussions, and offer a safe space to share new ideas.
See you at the next issue! 👋