Portal Weekly #38: Evo genomic foundation model, scaling ML docking, and more.
Join Portal and stay up to date on research happening at the intersection of tech x bio 🧬
Hi everyone 👋
Welcome to another issue of the Portal newsletter where we provide weekly updates on talks, events, and research in the TechBio space!
📅 TechBio Events on the Horizon
Nucleate is launching its Canadian chapter with an AI in BioTech event in Montreal on Wednesday, March 13th. Don’t miss out on an opportunity to hear from various panellists and meet more folks in the TechBio community! Sign up here.
For those of you based in London, Recursion and Valence Labs will be hosting a TechBio mixer in London on Tuesday, March 12th! DM the Valence Labs account to get more details about the event.
💬 Upcoming talks
M2D2 continues next week with a talk by Chenyu Wang, who will introduce an information maximization approach to deal with batch effects, which are a major difficulty that can lead to systematic errors and non-biological associations.
Join us live on Zoom on Tuesday, March 12th at 11 am ET. Find more details here.
LoGG continues with Zahra Kadkhodaie, who will demonstrate the effects of powerful inductive biases in deep neural network architectures and/or training algorithms, and will likely answer a lot of questions about generalization properties of diffusion models.
Join us live on Zoom on Monday, March 11th at 11 am ET. Find more details here, and the LoGG calendar here.
Speakers for CARE talks are usually announced on Fridays after this newsletter goes out. Sign up for Portal and get notifications when a new talk is announced on the CARE events page. You can also follow us on Twitter!
If you enjoy this newsletter, we’d appreciate it if you could forward it to some of your friends! 🙏
Let’s jump right in!👇
📚 Community Reads
LLMs for Science
L+M-24: Building a Dataset for Language + Molecules @ ACL 2024
Training (or at least fine-tuning) language models on domain-specific data usually helps to boost performance, but in the molecular domain, it’s hard to get molecule-language pair datasets for training. Datasets can be large but noisy and made by entity-linking on the scientific literature or by converting property prediction datasets to natural language using templates. This document explains the creation of a dataset for a workshop at ACL2024 (the association for computational linguistics). Designing a dataset with natural language principles in mind brings in knowledge from a different domain (linguistics) and will provide an interesting complement to existing dataset creation approaches: according to the authors, their dataset overall proves to be quite challenging.
ML for Small Molecules
Combining IC50 or Ki Values from Different Sources Is a Source of Significant Noise
ML approaches require a lot of data, especially deep learning models. Researchers often combine literature data to make large datasets, even though it’s well-known that this is scientifically risky for measurements like IC50. This study estimates the amount of noise present in combined cases, finding that a lot of assays selected using minimal curation have minimal agreement with each other. Although this seems like a study to which many would say “Well, of course! I expected that!”, it’s important to have actual quantification of these phenomena so that we can more clearly define the problem, and design better solutions.
A Resource to Enable Chemical Biology and Drug Discovery of WDR Proteins
WD40-repeat (WDR) proteins are one of the largest human protein families and include more essential genes in cancer than any other protein family, but are generally understudied. This study releases a range of materials, methods, new crystal structures, and assays on druggability for WDR proteins. They also do a hit-finding pilot, identifying drug-like ligands for 9 of 20 targets. The tools and resources provided here make it more likely that other researchers will try to tackle this class of proteins, which could help propel treatments for a range of diseases.
ML for Atomistic Simulations
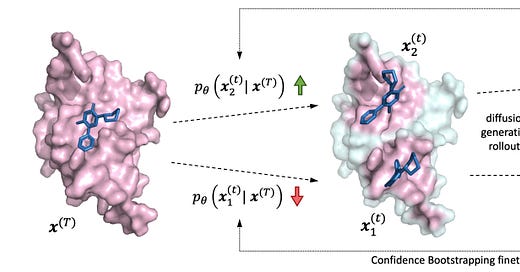
Deep Confident Steps to New Pockets: Strategies for Docking Generalization
ML-based molecular docking has promise for quickly telling us whether a molecule binds well to a protein, but we need to be careful to make sure our models can generalize well. This study presents a new benchmark based on the ligand-binding domains of proteins and suggests that existing ML-based docking models generalize poorly. They analyze docking scaling laws and improve both performance and generalization across benchmarks, and also introduce a new training paradigm that seems to improve how well ML-based methods can dock to unseen protein classes.
Interested to learn more about physics-based simulations? Check out the LoGG Talk with Bowen Jing where he presented the paper “AlphaFold Meets Flow Matching for Generating Protein Ensembles”.
Graph Learning
Multi-State RNA Design with Geometric Multi-Graph Neural Networks
RNA is not just a squiggly line as presented in many textbooks and blogs: it folds into 3D structures that can drive specific biological functions. Accompanying the paper linked above, a blog-style post and tutorial describe a geometric RNA design pipeline to design RNA backbone structures that explicitly account for 3D structure and dynamics by using an SE(3) equivariant encoder-decoder graph neural network framework. The tutorial has intuitive pictures of the model, and if you have any questions you can probably reach out to the author on Portal!
ML for Proteins
PTM-Mamba: A PTM-Aware Protein Language Model with Bidirectional Gated Mamba Blocks
Post-translational modifications (PTMs) can change how a protein acts, but crystal structures don’t often capture them. Protein language models don’t rely on crystal structures and could be useful predictors of PTMs. This study leverages ESM-2 embeddings that have already performed well and adds PTM information through unique token embeddings; although there aren’t many good PTM-specific benchmarks, this approach seems to improve on ESM-2 alone and more experiments and benchmarking efforts are ongoing. The authors used Mamba for its efficiency for a limited compute budget - they explained more of their reasoning in a Portal discussion on the paper!
ML for Omics
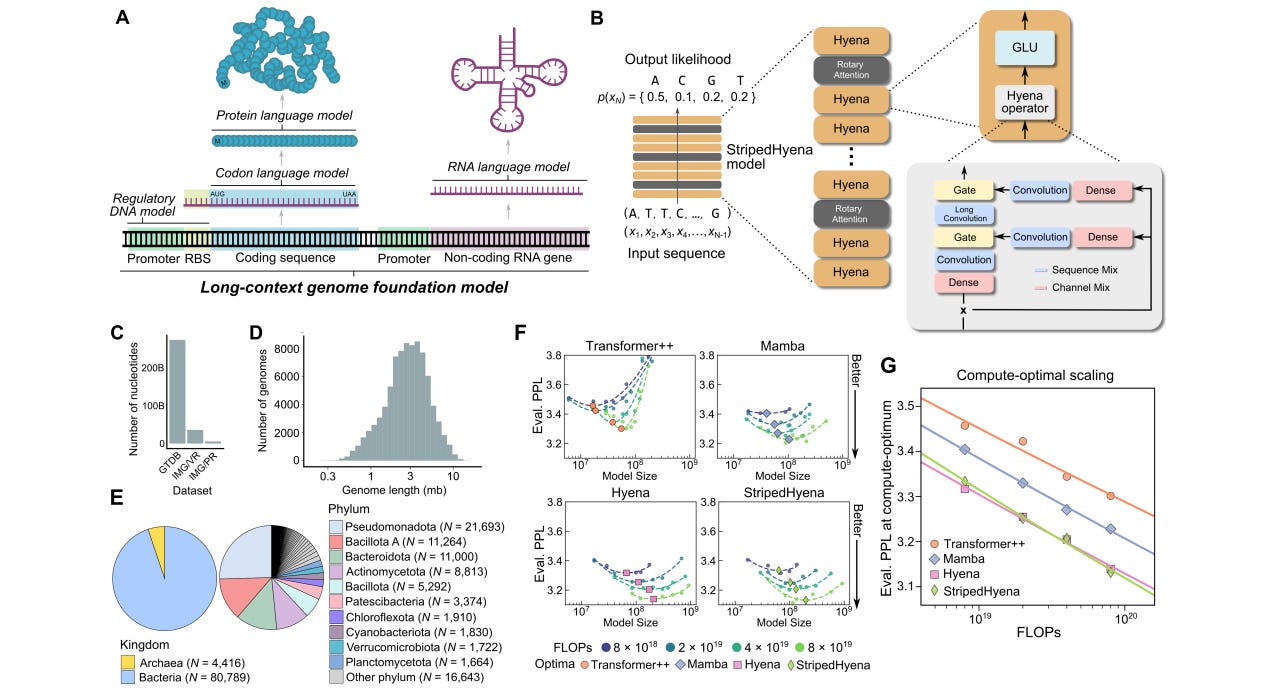
Sequence modeling and design from molecular to genome scale with Evo
Foundation models are supposed to learn broad patterns that can be applied over a range of tasks. This study presents a genomic foundation model trained on prokaryotic (bacteria) and virus genomes, leveraging StripedHyena architecture (a combination of Hyena and Transformer architectures) to try to improve scaling performance. The model is said to be able to generalize across DNA, RNA and protein, perform zero-shot function prediction comparable to domain-specific language models, and generate multiple elements.
If you want to learn more about genomic foundation models, watch the M2D2 talk from first author Eric Nguyen on HyenaDNA!
Think we missed something? Join our community to discuss these topics further!
🎬 Latest Recordings
LoGG
Manifold Diffusion Fields by Ahmed Elhag
CARE
Linear Structure of High-Level Concepts in Text-Controlled Generative Models by Victor Veitch
You can always catch up on previous recordings on our YouTube channel or the Portal Events page!
Portal is the home of the TechBio community. Join here and stay up to date on the latest research, expand your network, and ask questions. In Portal, you can access everything that the community has to offer in a single location:
M2D2 - a weekly reading group to discuss the latest research in AI for drug discovery
LoGG - a weekly reading group to discuss the latest research in graph learning
CARE - a weekly reading group to discuss the latest research in causality
Blogs - tutorials and blog posts written by and for the community
Discussions - jump into a topic of interest and share new ideas and perspectives
See you at the next issue! 👋