Portal Weekly #47: AlphaFold 3, a first analysis on AI-discovered drugs in clinical trials, and more.
Join Portal and stay up to date on research happening at the intersection of tech x bio 🧬
Hi everyone 👋
Welcome to another issue of the Portal newsletter where we provide weekly updates on talks, events, and research in the TechBio space!
We had an awesome time meeting so many community members at our TechBio social in Vienna for ICLR! To stay updated on future events and hear about them first, join Portal and follow us on X and LinkedIn. We’ll be doing more events like this in the future :)
💻 Latest Blogs
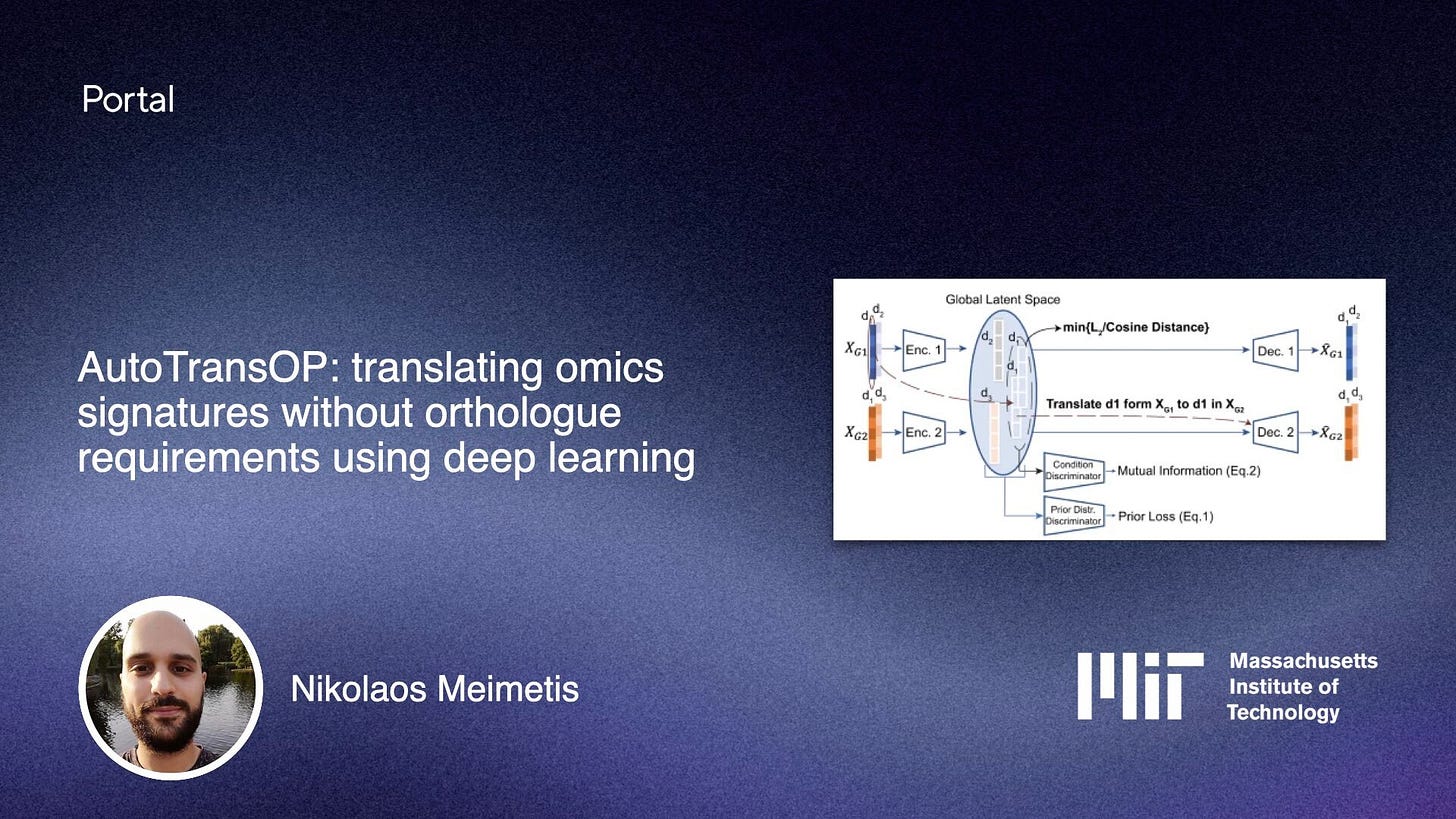
Clinical trials can fail despite promising results on animal models during development, partially because these models don’t fully capture human biology. In the latest blog post, Nikolaos Meimetis describes a deep learning framework to translate across biological contexts, without needing your omics profiles to have common biomolecules (orthologs). Check it out here!
Interested in sharing your work with the community? Fill out our interest form to get started.
💬 Upcoming talks
Have you heard about Kolmogorov-Arnold Networks yet? Next week at LoGG, we have Ziming Liu, first author of the paper, who will introduce this proposed drop-in replacement for multi-layer perceptrons (MLPs), that can increase interpretability and speed and may be particularly useful in math and physics for symbolic discovery.
Join us live on Zoom on Monday, April 22nd at 11 am ET. Find more details here.
Speakers for CARE talks are usually announced on Fridays after this newsletter goes out. Sign up for Portal and get notifications when a new talk is announced on the CARE events page. You can also follow us on Twitter and LinkedIn!
If you enjoy this newsletter, we’d appreciate it if you could forward it to some of your friends! 🙏
Let’s jump right in!👇
📚 Community Reads
LLMs for Science
Autonomous LLM-Driven Research from Data to Human-Verifiable Research Papers
Autonomous research systems are poised to unleash a new age of scientific discovery. With the goal of mimicking human scientific practices, this study takes interacting LLM agents through a complete stepwise research process, with a human in the loop for oversight. Although the novelty of the research was rather limited, the nice thing is that they say that their results are verifiable through information-tracing because of the sequential process.
ML for Small Molecules
Equivariant 3D-Conditional Diffusion Model for Molecular Linker Design
In fragment-based drug design (FBDD), we start with small molecular compounds (fragments) that interact with a target protein pocket and then try to combine them into a single chemical compound. This paper introduces a way of joining fragments placed in a 3D context while conditioning the design on a target protein pocket, independently generating molecules that match potent drugs reported in the literature.
De Novo Generation of Multi-Target Compounds Using Deep Generative Chemistry
Drugs with multiple targets have promise in treating diseases that are resistant to single-target drugs, like some forms of cancer. This study describes a model, POLYGON, that uses generative reinforcement learning to generate molecular structures two inhibit two protein targets. Synthesizing 32 compounds and testing them in the lab, they find >50% reduction in activity of each of their targets, supporting the potential of this approach.
ML for Proteins
Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3
AlphaFold has revolutionized protein structure prediction and spawned a whole ecosystem of approaches that leverage it in different ways. This week, Google DeepMind and Isomorphic announced AlphaFold3 in a blog and a Nature publication. With diffusion-based architecture, the new model can predict structures of complexes including proteins, nucleic acids, small molecules, ions, and modified residues. Unfortunately, interfacing with this model is currently limited to a web-server, with no code released; the model is restricted to no commercial use, use in building docking tools, or training ML models, which would limit what the community can do with it.
There’s already been some discussion about the model on Portal. Do you think this model will still be revolutionary if it stays closed-source? Head over and contribute your thoughts!
Predictomes: A Classifier-Curated Database of AlphaFold-Modeled Protein-Protein Interactions
One of the useful things that you can do with a more open-source AlphaFold is to use its structure predictions to investigate protein-protein interactions (PPIs). AlphaFold-Multimer can help predict PPIs but its confidence metrics don’t necessarily exclude false positives. This study trains a classifier to separate true and false PPIs, including in proteome-wide screens; they also generate an all-by-all matrix of ~300 human genome maintenance proteins, provided in a webserver. This study helps provide a framework for interpreting large-scale AF-M screens and increasing confidence in PPI predictions.
ML for Omics
State of the Interactomes: an Evaluation of Molecular Networks for Generating Biological Insights
The “interactome” is a nebulous concept; there are many types of biomolecules (proteins, RNA, DNA, lipids among others) that all interact with each other in different ways. This study launches an evaluation of 46 current human interactomes, showing that not all networks are created equal: large composite networks like STRING were most effective for identifying disease genes, and smaller networks like SIGNOR showed strong interaction prediction performance.
Open-Source
High-throughput Quantum Theory of Atoms in Molecules (QTAIM) for Geometric Deep Learning of Molecular and Reaction Properties
This paper presents a package for geometric molecular property prediction based on topological features of quantum mechanical electron density, with code that is compatible with BondNet and ChemProp architectures.
Reviews
How Successful are AI-Discovered Drugs in Clinical Trials? A First Analysis and Emerging Lessons
To realize the promise of AI for drug discovery, these molecules have to pass clinical trials. This review provides insight into the relative success of AI-discovered molecules, with one main finding that they have a higher Phase I success rate than historic industry averages.
Think we missed something? Join our community to discuss these topics further!
🎬 Latest Recordings
LoGG
CARE
You can always catch up on previous recordings on our YouTube channel or the Portal Events page!
Portal is the home of the TechBio community. Join here and stay up to date on the latest research, expand your network, and ask questions. In Portal, you can access everything that the community has to offer in a single location:
M2D2 - a weekly reading group to discuss the latest research in AI for drug discovery
LoGG - a weekly reading group to discuss the latest research in graph learning
CARE - a weekly reading group to discuss the latest research in causality
Blogs - tutorials and blog posts written by and for the community
Discussions - jump into a topic of interest and share new ideas and perspectives
See you at the next issue! 👋