Portal Weekly #54: new benchmarks for LLMs for science, a text2protein model, double-ended synthesis planning, and more.
Join Portal and stay up to date on research happening at the intersection of tech x bio 🧬
Hi everyone 👋
Welcome to another issue of the Portal newsletter where we provide weekly updates on talks, events, and research in the TechBio space!
📅 Upcoming Events
Polaris is a new benchmarking platform for ML in drug discovery. Don’t miss out on the Polaris launch event next Thursday at ICML!
The Polaris team is also giving out a Polaris merch box for anyone who uploads a dataset or benchmark before noon CET on July 25th. You can get early access to Polaris here and find more details about participating in the launch showcase here.
💻 Latest Blogs
Protein-protein interactions are not only about how structures interact - the strength of the interaction matters. In a new blog post, Wei Lu shows how the structure-predicting AlphaFold3 can contribute to an ensemble of binding-affinity prediction models. Check it out here!
If you enjoy this newsletter, we’d appreciate it if you could forward it to some of your friends! 🙏
Let’s jump right in!👇
📚 Community Reads
LLMs for Science
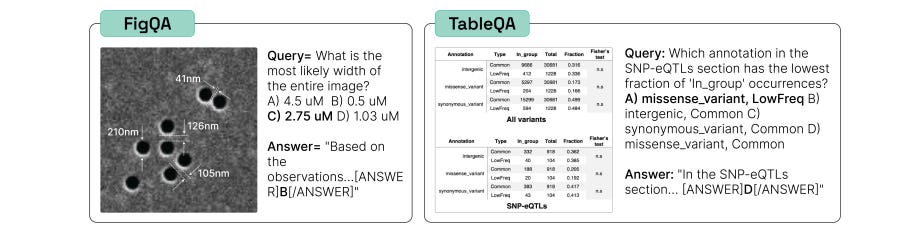
Two recent publications argue that benchmarks for LLMs need to evaluate their capabilities in realistic scientific research problems. SciCode: A Research Coding Benchmark Curated by Scientists presents challenging coding problems across physics, math, material science, biology, and chemistry, drawn from scripts that scientists use in everyday workflows. The best-performing LLM that they tested solved only ~5% of the problems in the most realistic setting, demonstrating that LLMs still have a way to go before being useful in many practical scientific coding tasks. LAB-Bench: Measuring Capabilities of Language Models for Biology Research proposes ~2500 multiple choice questions that cover practical biology research tasks representing early capabilities we would want an AI assistant for, including reasoning over literature, interpreting figures and tables, and manipulating DNA and protein sequences. Improving on either of these benchmarks would show that LLMs have significantly advanced in their usefulness for scientific pursuits.
SciCode: A Research Coding Benchmark Curated by Scientists
LAB-Bench: Measuring Capabilities of Language Models for Biology Research
ML for Small Molecules
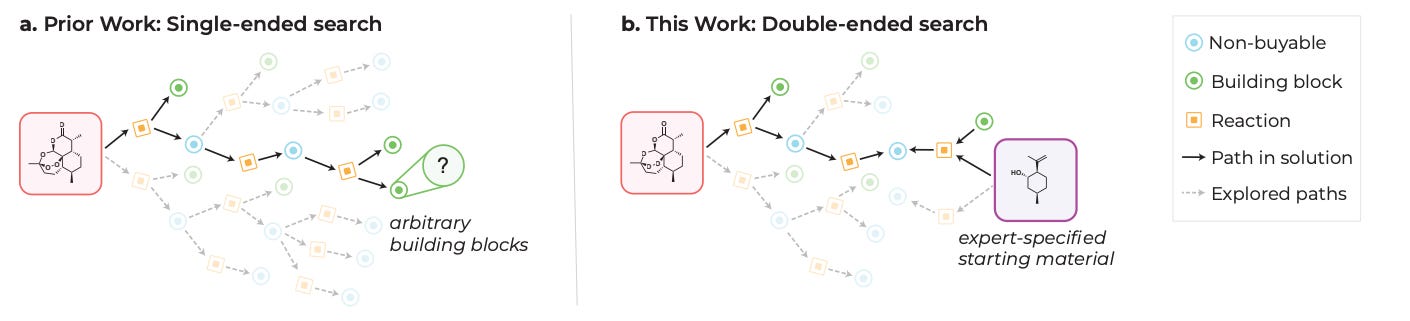
Double-Ended Synthesis Planning with Goal-Constrained Bidirectional Search
If you’re planning to synthesize some potentially great drug candidate, you start from the final product and work backwards to figure out your starting material (‘retrosynthesis’). Experienced chemists may start with specific starting materials in mind (‘structure-goals’), which helps to constrain the synthesis solution space to something more manageable. This study suggests a ‘bidirectional search’ algorithm that learns from expert synthesis trajectories, for a potentially more efficient retrosynthesis planner with better solve rates.
The Impact of Library Size and Scale of Testing on Virtual Screening
You can virtually screen through bigger and bigger libraries these days, but experimental testing doesn't scale as well so it's hard to actually say what is the actual hit rate of virtual screening. This study tried to look at both virtual library screening size and number of molecules tested by docking a 1.7 billion molecule virtual library against the model enzyme AmpC β-lactamase, testing 1,521 new molecules and comparing the results to the same screen with a library of 99 million molecules, where only 44 molecules were tested. Their results support the idea that there are many more compounds to be discovered than are being tested.
ML for Atomistic Simulations
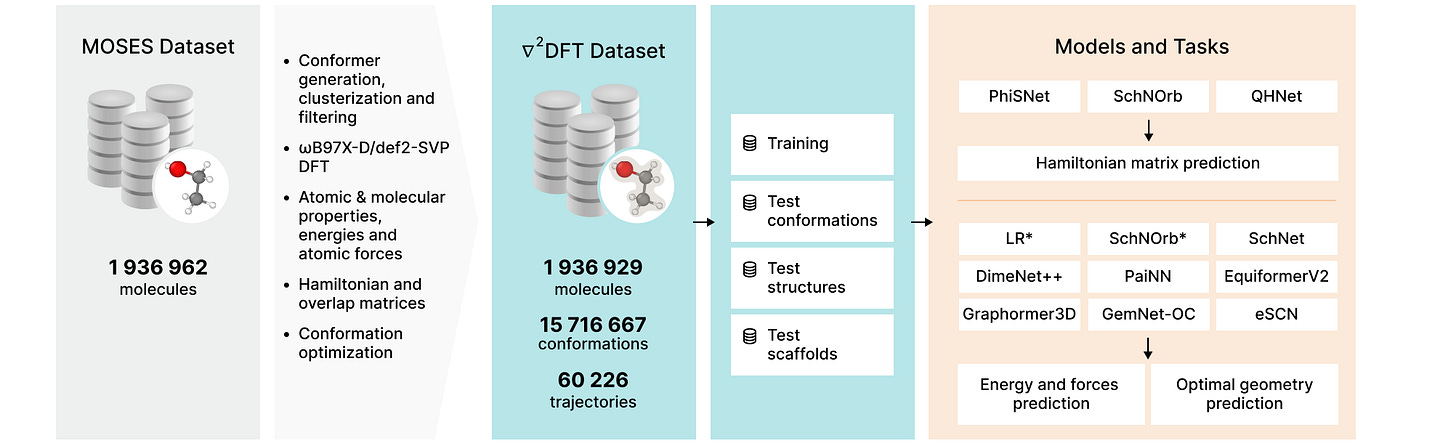
∇2DFT: A Universal Quantum Chemistry Dataset of Drug-Like Molecules and a Benchmark for Neural Network Potentials
Neural Network potentials can reduce the time and computational cost of determining molecular properties, but they need large datasets for training. This work presents a new dataset of almost 2 million molecules and >15 million conformations, including DFT-level calculations for each molecular conformation, as well as energies, forces, 17 molecular properties, Hamiltonian and overlap matrices, and a wavefunction object.
ML for Proteins
MPM4: AI Text2Protein Breakthrough Tackles the Molecule Programming Challenge
In a Medium post, 310 AI describes their latest text-to-protein model: the idea is that you can input a prompt like “This protein is involved in transporting other proteins within cells, specifically targeting them to vacuoles for various biological processes.” and get an AI-generated sequence back. The generated proteins are evaluated with three main metrics to evaluate sequence, structure, and function, and top ones supposedly approved by scientists. Currently ~1000 are deposited in a “MPM4 Repo” - take a look!
ML for Omics
Federated Learning for Predicting Compound Mechanism of Action Based on Image-Data from Cell Painting
Data is needed to train ML models, but especially in drug discovery little of the data is public: many companies keep their data internal and rarely disclose them. Federated Learning (FL) aims at training a joint model between multiple parties without disclosing data between the parties. This study evaluates how effective FL can be for predicting compound mechanisms of action using Cell Painting microscopy data, especially looking at the effects of data heterogeneity (differences across institutions). They suggest that FL can be beneficial for all concerned, and has potential for collaborative ML for drug discovery.
Open Source
DOPtools: a Python Platform for Descriptor Calculation and Model Optimization. Overview and Usage Guide
Many different tools and libraries calculate molecular descriptors, but the authors of this preprint argue that tools’ outputs need to be easily connected to standard ML libraries. This resource has a unified API for descriptors that can be fed as input into scikit-learn, and a command line interface for calculation of descriptors and eventual hyperparameters optimization of models.
Think we missed something? Join our community to discuss these topics further!
🎬 Latest Recordings
LoGG
G-Retriever: Retrieval-Augmented Generation for Textual Graph Understanding and Question Answering by Xiaoxin He
You can always catch up on previous recordings on our YouTube channel or the Portal Events page!
Portal is the home of the TechBio community. Join here and stay up to date on the latest research, expand your network, and ask questions. In Portal, you can access everything that the community has to offer in a single location:
M2D2 - a weekly reading group to discuss the latest research in AI for drug discovery
LoGG - a weekly reading group to discuss the latest research in graph learning
CARE - a weekly reading group to discuss the latest research in causality
Blogs - tutorials and blog posts written by and for the community
Discussions - jump into a topic of interest and share new ideas and perspectives
See you at the next issue! 👋